HW4 Show all work to receive
credit:
- A recent study compared the inhibition of COX-2 and COX-1 enzymes by the traditional NSAID's, aspirin and indomethacin, and a known selective COX-2 inhibitor, celebrex. The COX-1 and COX-2 enzyme activity was determined by measuring the PGE2 levels generated during the incubation of human platelets with the Amersham EIA assay. The IC50 of aspirin, indomethacin, and celebrex on COX-1 were 4.55 uM, 0.01 uM, and 1.4 uM, respectively. The IC50 of the test items on COX-2 were 595 uM, 0.4 uM, and 0.47 uM respectively.
- Graphically illustrate what is meant by the IC50 values
- Calculate the relative effectiveness of these drugs to inhibit COX-2 compared to COX-1 enzymes.
- On the same graph, plot the expected rate of PGE2 production vs Arachidonic Acid concentration with both no inhibitor present and with an inhibitor present.
- Clearly illustrate the noncovalent interactions expected between the two active site amino acid tyrosine and arginine side groups with aspirin by:
- For the arginine side group, show all bonds and appropriate charges at physiological pH; also draw the three important resonance structures
- Show all bonds and appropriate charges for aspirin at physiological pH
- Show the noncovalent interaction between arginine and aspirin; identify what type of interaction this is.
- Show the noncovalent interaction between tyrosine and aspirin; identify what type of interaction this is.
- While developing a new drug, you first characterize the chemical kinetics by measuring the velocity of the enzyme catalyzed reaction of interest with different substrate concentrations. The enzyme concentration is held constant at 10 nM in all experiments.
- Buffers are mixtures of weak acids and their conjugate bases (weak acid is structure with the proton; conjugate base is structure without the proton) and have concentration ratios in the 1:10 to 10:1 range to provide the best pH stability. Glutamic acid has three pKa's (see class handout or look them up) associated with it.
- Identify the three pH ranges over which glutamic acid could be used as a buffer.
- For each of these three ranges, show the structure (showing all bonds and charges) of the two most concentrated forms present.
- At physiological plasma pH, draw the two most prevalent structures
- Calculate the expected ratio of concentrations of the two most prevalent structures at physiological plasma pH.
- Draw the structure that is most prevalent at a stomach pH.
- Draw the reaction and associated mechanism for the hydrolysis of a phospholipid (having a choline) to release arachidonic acid. Show the molecular structure of all reactants and products.
[substrate] (mM) Initial Velocity (mmol/min)
---------------------------------------------------------------------
3.0 10.4
5.0 14.5
10.0 22.5
30.0 33.8
90.0 40.5
Use these data and a Lineweaver-Burke plot to obtain Vmax,
the turnover number, and KM for this enzyme-substrate combination.
- Draw the reaction associated with the formation of a dipeptide from serine and cysteine.
- For the peptide bond in the dipeptide just drawn, identify the expected hybridization of all C, N, and O atoms near this bond. Then draw the two peptide bond resonance structures and explain whether your answer to the hybridization question needs to be changed.
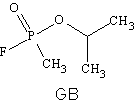
- The nerve agent GB reacts with a serine side chain at the acetylcholinesterase active site in a manner similar to how aspirin reacts with COX-1 enzymes. GB is an organophosphorous ester with the below structure. Clearly show how you would expect the serine R group to react with GB to form a covalent bond. Clearly show the structures of the expected products.