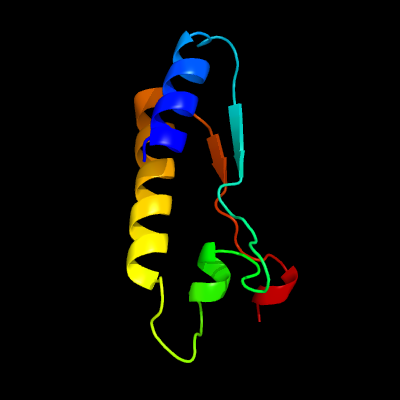
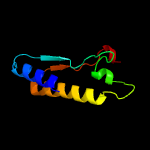
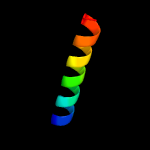
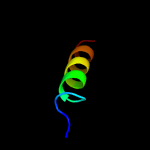
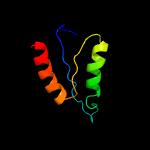
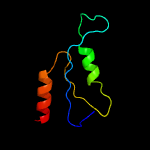
1 c5xczA_
48.5
28
PDB header: hydrolaseChain: A: PDB Molecule: glucanase;PDBTitle: structure of the cellobiohydrolase cel6a from phanerochaete2 chrysosporium in complex with cellobiose at 2.1 angstrom
2 c1kddC_
44.4
33
PDB header: de novo proteinChain: C: PDB Molecule: gcn4 acid base heterodimer acid-d12la16i;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16i base-d12la16l
3 c1kddF_
44.0
33
PDB header: de novo proteinChain: F: PDB Molecule: gcn4 acid base heterodimer acid-d12la16i;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16i base-d12la16l
4 c1kddA_
44.0
33
PDB header: de novo proteinChain: A: PDB Molecule: gcn4 acid base heterodimer acid-d12la16i;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16i base-d12la16l
5 c1kd9C_
42.1
33
PDB header: de novo proteinChain: C: PDB Molecule: gcn4 acid base heterodimer acid-d12la16l;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16l base-d12la16l
6 c1kd9F_
42.1
33
PDB header: de novo proteinChain: F: PDB Molecule: gcn4 acid base heterodimer acid-d12la16l;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16l base-d12la16l
7 c1kd9A_
42.1
33
PDB header: de novo proteinChain: A: PDB Molecule: gcn4 acid base heterodimer acid-d12la16l;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16l base-d12la16l
8 d1tzpa_
32.5
21
Fold: Hedgehog/DD-peptidaseSuperfamily: Hedgehog/DD-peptidaseFamily: MepA-like
9 c6nbhP_
29.6
33
PDB header: signaling proteinChain: P: PDB Molecule: long-acting parathyroid hormone analog;PDBTitle: cryo-em structure of parathyroid hormone receptor type 1 in complex2 with a long-acting parathyroid hormone analog and g protein
10 c2wb7B_
28.0
24
PDB header: unknown functionChain: B: PDB Molecule: pt26-6p;PDBTitle: pt26-6p
11 c2n8oA_
21.7
33
PDB header: antimicrobial proteinChain: A: PDB Molecule: bacteriocin aureocin a53;PDBTitle: nmr solution structure of aureocin a53
12 c3f29A_
21.7
19
PDB header: oxidoreductaseChain: A: PDB Molecule: eight-heme nitrite reductase;PDBTitle: structure of the thioalkalivibrio nitratireducens cytochrome c nitrite2 reductase in complex with sulfite
13 c1kd8F_
20.7
29
PDB header: de novo proteinChain: F: PDB Molecule: gcn4 acid base heterodimer acid-d12ia16v;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12ia16v base-d12la16l
14 c1kd8A_
20.7
29
PDB header: de novo proteinChain: A: PDB Molecule: gcn4 acid base heterodimer acid-d12ia16v;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12ia16v base-d12la16l
15 c1kd8C_
20.7
29
PDB header: de novo proteinChain: C: PDB Molecule: gcn4 acid base heterodimer acid-d12ia16v;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12ia16v base-d12la16l
16 c2yqpA_
19.8
47
PDB header: gene regulation, hydrolaseChain: A: PDB Molecule: probable atp-dependent rna helicase ddx59;PDBTitle: solution structure of the zf-hit domain in dead (asp-glu-2 ala-asp) box polypeptide 59
17 c3h3gB_
19.7
42
PDB header: membrane proteinChain: B: PDB Molecule: parathyroid hormone-related protein;PDBTitle: crystal structure of the extracellular domain of the human parathyroid2 hormone receptor (pth1r) in complex with parathyroid hormone-related3 protein (pthrp)
18 c3orsD_
19.3
23
PDB header: isomerase,biosynthetic proteinChain: D: PDB Molecule: n5-carboxyaminoimidazole ribonucleotide mutase;PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide mutase2 from staphylococcus aureus
19 c2ywxA_
18.1
30
PDB header: lyaseChain: A: PDB Molecule: phosphoribosylaminoimidazole carboxylase catalytic subunit;PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase2 catalytic subunit from methanocaldococcus jannaschii
20 d2ewca1
15.3
11
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: YjgF/L-PSP
21 c3wdkA_
not modelled
15.1
65
PDB header: ligaseChain: A: PDB Molecule: 4-phosphopantoate--beta-alanine ligase;PDBTitle: crystal structure of 4-phosphopantoate-beta-alanine ligase complexed2 with reaction intermediate
22 c3vohA_
not modelled
15.1
26
PDB header: hydrolaseChain: A: PDB Molecule: cellobiohydrolase;PDBTitle: cccel6a catalytic domain complexed with cellobiose
23 c3lp6D_
not modelled
15.0
27
PDB header: lyaseChain: D: PDB Molecule: phosphoribosylaminoimidazole carboxylase catalytic subunit;PDBTitle: crystal structure of rv3275c-e60a from mycobacterium tuberculosis at2 1.7a resolution
24 d1o4va_
not modelled
14.7
27
Fold: Flavodoxin-likeSuperfamily: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)Family: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
25 c4kueA_
not modelled
14.6
23
PDB header: oxidoreductaseChain: A: PDB Molecule: 3-hydroxybutyryl-coa dehydrogenase;PDBTitle: crystal structure of 3-hydroxybutylryl-coa dehydrogenase from2 clostridium butyricum
26 c3trhI_
not modelled
14.5
25
PDB header: lyaseChain: I: PDB Molecule: phosphoribosylaminoimidazole carboxylasePDBTitle: structure of a phosphoribosylaminoimidazole carboxylase catalytic2 subunit (pure) from coxiella burnetii
27 d3gata_
not modelled
13.3
35
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Erythroid transcription factor GATA-1
28 c4kw2A_
not modelled
13.2
16
PDB header: isomeraseChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative uncharacterized protein (bdi_1873)2 from parabacteroides distasonis atcc 8503 at 2.32 a resolution
29 c6qb7A_
not modelled
13.0
27
PDB header: signaling proteinChain: A: PDB Molecule: btb/poz domain-containing protein kctd16;PDBTitle: structure of the h1 domain of human kctd16
30 c2kaeA_
not modelled
13.0
17
PDB header: transcription/dnaChain: A: PDB Molecule: gata-type transcription factor;PDBTitle: data-driven model of med1:dna complex
31 d1qjwa_
not modelled
12.9
26
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycosyl hydrolases family 6, cellulasesFamily: Glycosyl hydrolases family 6, cellulases
32 c3couA_
not modelled
12.7
41
PDB header: hydrolaseChain: A: PDB Molecule: nucleoside diphosphate-linked moiety x motif 16;PDBTitle: crystal structure of human nudix motif 16 (nudt16)
33 c2l5gB_
not modelled
12.7
33
PDB header: transcription regulatorChain: B: PDB Molecule: putative uncharacterized protein ncor2;PDBTitle: co-ordinates and 1h, 13c and 15n chemical shift assignments for the2 complex of gps2 53-90 and smrt 167-207
34 c3he5D_
not modelled
12.1
38
PDB header: de novo proteinChain: D: PDB Molecule: synzip2;PDBTitle: heterospecific coiled-coil pair synzip2:synzip1
35 c3ffdP_
not modelled
12.0
36
PDB header: immune system/hormoneChain: P: PDB Molecule: parathyroid hormone-related protein;PDBTitle: structure of parathyroid hormone-related protein complexed to a2 neutralizing monoclonal antibody
36 c3l3fX_
not modelled
11.7
37
PDB header: protein bindingChain: X: PDB Molecule: protein doa1;PDBTitle: crystal structure of a pfu-pul domain pair of saccharomyces cerevisiae2 doa1/ufd3
37 d2fzva1
not modelled
11.7
23
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: NADPH-dependent FMN reductase
38 c6o55B_
not modelled
11.5
21
PDB header: isomeraseChain: B: PDB Molecule: n5-carboxyaminoimidazole ribonucleotide mutase;PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide mutase2 (pure) from legionella pneumophila
39 d1nc5a_
not modelled
11.2
15
Fold: alpha/alpha toroidSuperfamily: Six-hairpin glycosidasesFamily: Hypothetical protein YteR
40 c6n7rI_
not modelled
11.1
26
PDB header: rna binding proteinChain: I: PDB Molecule: protein luc7;PDBTitle: saccharomyces cerevisiae spliceosomal e complex (act1)
41 d1gnfa_
not modelled
10.8
50
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Erythroid transcription factor GATA-1
42 d1y0ja1
not modelled
10.7
50
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Erythroid transcription factor GATA-1
43 c4b4kK_
not modelled
10.6
26
PDB header: isomeraseChain: K: PDB Molecule: n5-carboxyaminoimidazole ribonucleotide mutase;PDBTitle: crystal structure of bacillus anthracis pure
44 c6s9kB_
not modelled
10.5
60
PDB header: signaling proteinChain: B: PDB Molecule: caspase-2;PDBTitle: structure of 14-3-3 gamma in complex with caspase-2 peptide containing2 14-3-3 binding motif ser139 and nls
45 c1emzA_
not modelled
10.5
78
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein e1;PDBTitle: solution structure of fragment (350-370) of the2 transmembrane domain of hepatitis c envelope glycoprotein3 e1
46 d2vuti1
not modelled
10.5
50
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Erythroid transcription factor GATA-1
47 c4mybA_
not modelled
10.5
40
PDB header: transferaseChain: A: PDB Molecule: 2-c-methyl-d-erythritol 4-phosphate cytidylyltransferase;PDBTitle: crystal structure of francisella tularensis 2-c-methyl-d-erythritol 4-2 phosphate cytidylyltransferase (ispd)
48 d1dota2
not modelled
10.2
31
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Transferrin
49 d5gata_
not modelled
10.0
50
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Erythroid transcription factor GATA-1
50 c3p6lA_
not modelled
9.8
10
PDB header: isomeraseChain: A: PDB Molecule: sugar phosphate isomerase/epimerase;PDBTitle: crystal structure of a sugar phosphate isomerase/epimerase (bdi_1903)2 from parabacteroides distasonis atcc 8503 at 1.85 a resolution
51 d1xmpa_
not modelled
9.8
26
Fold: Flavodoxin-likeSuperfamily: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)Family: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
52 c7ns4b_
not modelled
9.5
13
PDB header: ligaseChain: B: PDB Molecule: PDBTitle: catalytic module of yeast chelator-gid sr4 e3 ubiquitin ligase
53 c3ctvA_
not modelled
9.4
20
PDB header: oxidoreductaseChain: A: PDB Molecule: 3-hydroxyacyl-coa dehydrogenase;PDBTitle: crystal structure of central domain of 3-hydroxyacyl-coa dehydrogenase2 from archaeoglobus fulgidus
54 c6r0xE_
not modelled
9.3
35
PDB header: blood clottingChain: E: PDB Molecule: megakaryocyte and platelet inhibitory receptor g6b;PDBTitle: the extracellular domain of g6b-b in complex with fab fragment and2 dp12 heparin oligosaccharide.
55 c3cf4A_
not modelled
9.2
25
PDB header: oxidoreductaseChain: A: PDB Molecule: acetyl-coa decarboxylase/synthase alpha subunit;PDBTitle: structure of the codh component of the m. barkeri acds complex
56 c1r1gB_
not modelled
9.1
67
PDB header: toxinChain: B: PDB Molecule: neurotoxin bmk37;PDBTitle: crystal structure of the scorpion toxin bmbkttx1
57 d1mtyd_
not modelled
8.8
24
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ribonucleotide reductase-like
58 d1i52a_
not modelled
8.6
58
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: Cytidylytransferase
59 c6bv7A_
not modelled
8.6
18
PDB header: membrane proteinChain: A: PDB Molecule: sodium/calcium exchanger 1;PDBTitle: nmr structure of sodium/calcium exchanger 1 (ncx1) two-helix bundle2 (thb) domain
60 c4grdA_
not modelled
8.6
25
PDB header: lyase,isomeraseChain: A: PDB Molecule: phosphoribosylaminoimidazole carboxylase catalytic subunit;PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase2 catalytic subunit from burkholderia cenocepacia j2315
61 c2v4xA_
not modelled
8.4
25
PDB header: viral proteinChain: A: PDB Molecule: capsid protein p27;PDBTitle: crystal structure of jaagsiekte sheep retrovirus capsid n-2 terminal domain
62 c6sifG_
not modelled
8.3
8
PDB header: structural proteinChain: G: PDB Molecule: epidermicin locus structural protein;PDBTitle: epidermicin antimicrobial protein from staphylococcus epidermidis
63 c2k89A_
not modelled
8.3
55
PDB header: protein bindingChain: A: PDB Molecule: phospholipase a-2-activating protein;PDBTitle: solution structure of a novel ubiquitin-binding domain from2 human plaa (pfuc, gly76-pro77 cis isomer)
64 c1r1gA_
not modelled
8.2
67
PDB header: toxinChain: A: PDB Molecule: neurotoxin bmk37;PDBTitle: crystal structure of the scorpion toxin bmbkttx1
65 d1r1ga_
not modelled
8.2
67
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Short-chain scorpion toxins
66 d2hq4a1
not modelled
8.1
38
Fold: PH1570-likeSuperfamily: PH1570-likeFamily: PH1570-like
67 d1u11a_
not modelled
7.9
23
Fold: Flavodoxin-likeSuperfamily: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)Family: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
68 c6m8sM_
not modelled
7.9
32
PDB header: signaling proteinChain: M: PDB Molecule: btb/poz domain-containing protein kctd12;PDBTitle: crystal structure of the kctd12 h1 domain in complex with gbeta1gamma22 subunits
69 d2enda_
not modelled
7.7
8
Fold: T4 endonuclease VSuperfamily: T4 endonuclease VFamily: T4 endonuclease V
70 c6mjeF_
not modelled
7.6
33
PDB header: cell cycleChain: F: PDB Molecule: dsn1p;PDBTitle: structure of candida glabrata csm1: s. cerevisiae dsn1 complex
71 c6mjeB_
not modelled
7.6
33
PDB header: cell cycleChain: B: PDB Molecule: dsn1p;PDBTitle: structure of candida glabrata csm1: s. cerevisiae dsn1 complex
72 c6mjeH_
not modelled
7.6
33
PDB header: cell cycleChain: H: PDB Molecule: dsn1p;PDBTitle: structure of candida glabrata csm1: s. cerevisiae dsn1 complex
73 c6mjeD_
not modelled
7.6
33
PDB header: cell cycleChain: D: PDB Molecule: dsn1p;PDBTitle: structure of candida glabrata csm1: s. cerevisiae dsn1 complex
74 d1qcza_
not modelled
7.6
25
Fold: Flavodoxin-likeSuperfamily: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)Family: N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
75 c4ky3A_
not modelled
7.5
33
PDB header: de novo proteinChain: A: PDB Molecule: designed protein or327;PDBTitle: three-dimensional structure of the orthorhombic crystal of2 computationally designed insertion domain , northeast structural3 genomics consortium (nesg) target or327
76 c1kddD_
not modelled
7.5
25
PDB header: de novo proteinChain: D: PDB Molecule: gcn4 acid base heterodimer base-d12la16l;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16i base-d12la16l
77 c3pstA_
not modelled
7.3
32
PDB header: nuclear proteinChain: A: PDB Molecule: protein doa1;PDBTitle: crystal structure of pul and pfu(mutate) domain
78 c2n8pA_
not modelled
7.2
17
PDB header: antimicrobial proteinChain: A: PDB Molecule: lacticin q;PDBTitle: solution structure of lacticin q
79 c3n41A_
not modelled
7.2
40
PDB header: viral proteinChain: A: PDB Molecule: e3 envelope glycoprotein;PDBTitle: crystal structure of the mature envelope glycoprotein complex2 (spontaneous cleavage) of chikungunya virus.
80 c4r0rA_
not modelled
7.2
25
PDB header: biosynthetic proteinChain: A: PDB Molecule: eboizn21;PDBTitle: ebolavirus gp prehairpin intermediate mimic
81 c3rggD_
not modelled
7.2
12
PDB header: lyaseChain: D: PDB Molecule: phosphoribosylaminoimidazole carboxylase, pure protein;PDBTitle: crystal structure of treponema denticola pure bound to air
82 c3iufA_
not modelled
7.2
50
PDB header: protein bindingChain: A: PDB Molecule: zinc finger protein ubi-d4;PDBTitle: crystal structure of the c2h2-type zinc finger domain of human ubi-d4
83 c2fw9A_
not modelled
7.2
23
PDB header: lyaseChain: A: PDB Molecule: n5-carboxyaminoimidazole ribonucleotide mutase;PDBTitle: structure of pure (n5-carboxyaminoimidazole ribonucleotide mutase)2 h59f from the acidophilic bacterium acetobacter aceti, at ph 8
84 c2i5oA_
not modelled
7.2
41
PDB header: transferaseChain: A: PDB Molecule: dna polymerase eta;PDBTitle: solution structure of the ubiquitin-binding zinc finger2 (ubz) domain of the human dna y-polymerase eta
85 d2uube2
not modelled
7.2
33
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Ribosomal S5 protein, N-terminal domain
86 d1mhyd_
not modelled
7.0
21
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ribonucleotide reductase-like
87 c4zbzA_
not modelled
7.0
17
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: family 4 uracil-dna glycosylase from sulfolobus tokodaii (free form,2 x-ray wavelength=1.5418)
88 c1p6gE_
not modelled
7.0
50
PDB header: ribosomeChain: E: PDB Molecule: 30s ribosomal protein s5;PDBTitle: real space refined coordinates of the 30s subunit fitted into the low2 resolution cryo-em map of the ef-g.gtp state of e. coli 70s ribosome
89 d1pkpa2
not modelled
6.8
50
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Ribosomal S5 protein, N-terminal domain
90 c5ym0A_
not modelled
6.8
32
PDB header: lyaseChain: A: PDB Molecule: dihydroxy-acid dehydratase, chloroplastic;PDBTitle: the crystal structure of dhad
91 c1kd8E_
not modelled
6.7
25
PDB header: de novo proteinChain: E: PDB Molecule: gcn4 acid base heterodimer base-d12la16l;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12ia16v base-d12la16l
92 c1kddE_
not modelled
6.7
25
PDB header: de novo proteinChain: E: PDB Molecule: gcn4 acid base heterodimer base-d12la16l;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16i base-d12la16l
93 c1kd9D_
not modelled
6.7
25
PDB header: de novo proteinChain: D: PDB Molecule: gcn4 acid base heterodimer base-d12la16l;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16l base-d12la16l
94 c1kd9E_
not modelled
6.7
25
PDB header: de novo proteinChain: E: PDB Molecule: gcn4 acid base heterodimer base-d12la16l;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16l base-d12la16l
95 d1r7aa1
not modelled
6.7
75
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain
96 c4b4fB_
not modelled
6.6
30
PDB header: hydrolaseChain: B: PDB Molecule: beta-1,4-exocellulase;PDBTitle: thermobifida fusca cel6b(e3) co-crystallized with cellobiose
97 c1kd8B_
not modelled
6.6
25
PDB header: de novo proteinChain: B: PDB Molecule: gcn4 acid base heterodimer base-d12la16l;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12ia16v base-d12la16l
98 c1kd8D_
not modelled
6.6
25
PDB header: de novo proteinChain: D: PDB Molecule: gcn4 acid base heterodimer base-d12la16l;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12ia16v base-d12la16l
99 c1kddB_
not modelled
6.6
25
PDB header: de novo proteinChain: B: PDB Molecule: gcn4 acid base heterodimer base-d12la16l;PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12la16i base-d12la16l